FROM EDITOR
CURRENT REVIEW
What is already known on this topic?
The Role of H. pylori: Helicobacter pylori is a highly prevalent infection and a primary cause of chronic gastritis, peptic ulcer disease, and gastric cancer. It is classified as a Group I carcinogen.
The Process of Carcinogenesis: The long-term persistence of H. pylori initiates a cascade of pathological changes in the gastric mucosa—from inflammation to atrophy, metaplasia, dysplasia, and ultimately carcinoma (the Correa cascade).
The Importance of Eradication: Timely eradication of H. pylori is an effective measure for the primary prevention of gastric cancer, capable of preventing or reversing precancerous lesions.
The Role of PPIs: Proton pump inhibitors (PPIs) are a key component of eradication therapy, enhancing the efficacy of co-administered antibiotics.
The Impact of Pharmacogenetics: The metabolism of most PPIs (e.g., omeprazole, lansoprazole, pantoprazole) is dependent on the cytochrome P450 enzyme CYP2C19. Genetic polymorphisms of the CYP2C19 gene determine the rate of their metabolism and, consequently, their therapeutic efficacy.
What does this article add?
Focus on the Russian Population: The article highlights data indicating a high prevalence of the CYP2C1917 allele (conferring increased enzyme function) within the Russian population. This suggests a predisposition to suboptimal efficacy of standard PPI doses in a substantial proportion of patients.
Comparative Analysis of PPIs: It emphasises that rabeprazole is the least affected by CYP2C19 polymorphism, whereas the pharmacokinetics and efficacy of omeprazole, lansoprazole, and pantoprazole are significantly influenced.
Specific Recommendations: The article discusses international guidelines (e.g., from the Dutch Pharmacogenetics Working Group - DPWG) that recommend dose escalation for specific PPIs in patients who are rapid (RM) or ultra-rapid (UM) metabolisers.
A Case for Personalisation: It synthesises evidence supporting the use of CYP2C19 pharmacogenetic testing (PGT) as a practical tool for overcoming treatment failure, particularly in populations with a high frequency of rapid metaboliser phenotypes.
How might this influence clinical practice in the foreseeable future?
Integration of PGT into Routine Care: Pharmacogenetic testing for CYP2C19 polymorphisms could become a standard pre-therapeutic assessment prior to initiating eradication therapy, especially in regions with a high prevalence of the *17 allele, such as Russia.
Personalised PPI Selection and Dosing:
For patients with RM/UM phenotypes, the rationale would be established for either prescribing higher doses of first-generation PPIs or, more favourably, selecting rabeprazole or esomeprazole due to their lower dependency on CYP2C19.
For patients with IM/PM phenotypes, standard PPI doses would be confirmed as adequate.
Improved Eradication Efficacy: This personalised approach would facilitate the selection of maximally effective first-line therapy, thereby reducing the risk of treatment failure, the development of antibiotic resistance, and the progression of precancerous conditions.
Evolution of Clinical Guidelines: National clinical guidelines may be updated to include sections on pharmacogenetics to optimise anti-Helicobacter therapy regimens.
Background. Helicobacter pylori infection is a major risk factor for gastric cancer, and its eradication is considered a primary preventive measure. Proton pump inhibitors (PPIs) are a cornerstone of eradication therapy, but their efficacy is significantly influenced by genetic polymorphisms in the CYP2C19 enzyme, which is responsible for their metabolism.
Objective. To summarize and present current research on the impact of CYP2C19 genetic polymorphism on the effectiveness of H. pylori eradication therapy.
Materials and methods. A literature review was conducted using Russian and international databases (RSCI, PubMed, ResearchGate) with keywords including "CYP2C19 polymorphism," "proton pump inhibitor metabolism," and "Helicobacter pylori eradication." A total of 41 publications most relevant to the topic were analyzed.
Results. The metabolism of first-generation PPIs (omeprazole, lansoprazole, pantoprazole) is highly dependent on CYP2C19 activity. Patients are classified into different metabolic phenotypes (ultrarapid – UM, rapid – RM, normal – NM, intermediate – IM, poor – PM) based on their CYP2C19 genotype. Evidence, primarily from Asian populations, indicates that NM and RM/UM phenotypes are associated with lower eradication rates due to accelerated PPI metabolism and reduced drug exposure, whereas IM and PM phenotypes show higher efficacy. The Russian population has a high frequency of the rapid metabolizer allele CYP2C19*17, suggesting potential suboptimal response to standard PPI doses. Rabeprazole and esomeprazole demonstrate less dependence on CYP2C19, leading to more consistent efficacy across different genotypes. Clinical guidelines (e.g., CPIC, DPWG) recommend genotype-guided PPI dosing to optimize therapy.
Conclusion. CYP2C19 genetic polymorphism is a critical determinant of PPI pharmacokinetics and the effectiveness of H. pylori eradication. Pharmacogenetic testing for CYP2C19 can be a valuable tool for personalizing anti-Helicobacter therapy, particularly in populations with a high prevalence of rapid metabolizer alleles, by enabling the selection of the most appropriate PPI and its dose to overcome refractoriness and improve treatment outcomes.
CLINICAL PHARMACOGENETICS
What is already known on this topic?
The toxicity of high-dose methotrexate (MTX) in the treatment of childhood acute lymphoblastic leukemia (ALL) is a significant and relevant clinical problem.
The development of toxicity is multifactorial and demonstrates considerable interindividual variability.
The role of transporter proteins, encoded by the ABCB1 and SLCO1B1 genes, in the pharmacokinetics and toxicity of MTX has been established in previous studies.
Polymorphisms in these genes have been associated with altered MTX concentrations and an increased risk of adverse drug reactions (e.g., neutropenia, mucositis).
What does this study add?
This study confirms the association of specific polymorphisms with the MTX safety profile in a Russian pediatric ALL population:
ABCB1 rs1128503 (CC) is associated with an increased risk of oropharyngeal mucositis.
SLCO1B1 rs4149056 (TT) is associated with an increased risk of infectious complications.
ABCB1 rs1045642 (TT) showed a potential association with neuro- and nephrotoxicity (requiring further investigation).
It reveals that severe hepatotoxicity, hematological toxicity, and mucositis significantly increase the frequency of infectious complications.
It highlights that the results are sensitive to the statistical correction methods used (Bonferroni vs. FDR), which is crucial for interpreting pharmacogenetic studies.
How might this influence clinical practice in the foreseeable future?
The findings justify the need for large-scale pharmacogenetic testing prior to the implementation of personalized approaches in routine clinical practice.
In the future, preemptive genotyping for ABCB1 and SLCO1B1 polymorphisms could help identify a subgroup of patients at high risk for severe adverse reactions (mucositis, infections).
This would enable more intensive monitoring, timely prophylaxis, or therapy adjustments for these high-risk patients, potentially improving overall treatment safety.
To enhance prediction accuracy, further research is needed that incorporates haplotype analysis, covariates, and genes involved in MTX metabolism.
Background. Methotrexate (MTX) in high doses (1–5 g/m2) is a key component of treatment protocols for acute lymphoblastic leukemia (ALL) in children. Interindividual variability in MTX toxicity is a crucial area of research aimed at enhancing the safety of therapy while maintaining its effectiveness.
Objective. To evaluate the role of polymorphisms of genes ABCB1 (C3435T, C1236T, 2677G>T/A, rs4148738c>T), SLCO1B1 T521C on the safety profile of methotrexate in children with ALL.
Materials and methods. The study is involved 124 patients with a confirmed diagnosis of ALL (C91.0 according to ICD-10) who underwent high-dose methotrexate treatment (greater than 1 g/m2). The severity of adverse reactions (ARs) was assessed using laboratory methods based on the National Cancer Institute's toxicity criteria (CTCAE v5.0 2018). The carriage of polymorphic variants was determined using allele-specific polymerase chain reaction (PCR) in real time. The results were statistically analyzed using the SPSS Statistics 26.0 software (USA).
Results. The safety analysis of high-dose MTX therapy revealed that the ABCB1 1236C>T polymorphism is a significant predictor of oropharyngeal mucositis during MTX treatment, with a higher risk for CC homozygotes. Patients with the TT genotype of the SLCO1B1 T521C rs4149056 gene have a 2.7-fold increased risk of severe infectious complications, while patients with the TT genotype of the ABCB1 C3435T gene have an elevated risk of nephrotoxicity (p = 0.035, OR: 8.3 (95 % CI: 0.83–82.2) and neurotoxicity (p = 0.041, OR: 2.3 (95 % CI: 1.02–5.12).
Conclusion. The results of the safety analysis of high-dose MTX therapy indicate the need for comprehensive pharmacogenetic testing before implementing this treatment in clinical practice.
What is already known on this topic?
The Treatment Challenge: Treating multidrug-resistant (MDR) and extensively drug-resistant (XDR) tuberculosis remains a significant challenge, with treatment success rates (54% in Russia) still far below the WHO target (80%).
The Role of Pharmacogenetics: The response to therapy depends on both phenotypic factors (age, comorbidities, etc.) and genetic factors affecting drug metabolism and transport.
Lack of Data for MDR/XDR-TB: While some pharmacogenetic markers are known for drug-susceptible TB (e.g., NAT2 for isoniazid), personalized approaches for modern MDR/XDR-TB regimens are underdeveloped, and relevant biomarkers are largely unstudied.
What does this study add?
Identifies Novel Efficacy Markers: This clinical study identified, for the first time, two specific genetic polymorphisms significantly associated with poor treatment outcomes in MDR/XDR-TB patients:
Homozygous genotype AA in the CYP3A5 gene (rs776746).
Homozygous genotype AA in the ABCG2 gene (rs2231142).
Identifies Safety/Toxicity Markers: Discovered polymorphisms associated with the risk of specific adverse drug reactions (ADRs):
Neurotoxicity: associated with the wild-type GG genotype in the ABCB1 gene (rs2032582).
Gastrointestinal reactions: associated with the homozygous TT genotype in the ABCB1 gene (rs1128503).
Protective Effect: A polymorphism in the SLCO1B1 gene (rs4149056) was found to reduce the risk of arthralgia.
Proposes a Pharmacogenetic Panel: Defined a specific set of genes (CYP3A5, ABCG2, ABCB1, SLCO1B1) whose analysis could be useful for predicting treatment outcomes.
How might this influence clinical practice in the foreseeable future?
Therapy Personalization: If confirmed in larger studies, genetic testing could allow clinicians to identify patients at high risk of treatment failure (carriers of CYP3A5 AA and ABCG2 AA) before starting therapy.
Tailored Treatment Strategies: For these high-risk patients, longer or intensified chemotherapy regimens could be planned from the outset to improve the chances of success.
Adverse Reaction Prevention: Identifying patients with a genetic risk for specific ADRs (e.g., neurotoxicity) would enable targeted monitoring and allow for proactive dose adjustments or preventive measures, improving treatment tolerability.
Guideline Development: This work lays the foundation for developing clinical guidelines on the use of pharmacogenetic testing in patients with MDR/XDR-TB.
Background. Treatment of patients with tuberculosis (TB) with multidrug-resistant (MDR) causative agent is often complicated by adverse reactions (AR) with forced drug discontinuation, its effectiveness is far from the target indicators and depends on a number of factors, including the patient's genetic characteristics. Pharmacogenetic markers of MDR-TB have not been studied; it is expected that their identification will improve the results of treatment based on a personalized approach.
Objective. to determine the pharmacogenetic markers associated with the efficacy and safety of treatment of patients with MDR TB.
Methods. A prospective cohort study included 40 patients with MDR-TB without HIV infection who received therapy with bedaquiline, linezolid, and a fluoroquinolone in 2023–2024. All patients had 3–5 ml of venous blood collected once, regardless of the duration of therapy. Real-time PCR was used to determine the presence of single-nucleotide polymorphisms in the genes for cytochromes (CYP3A4, CYP3A5), P-glycoprotein (ABCB1), the membranebound ATP-binding cassette transporter (ABCG2), and the organic anion transporter (SLCO1B1), which were selected based on literature analysis and the PharmGKB database. The relationship between these indicators and the effectiveness and safety of treatment was assessed using univariate analysis, with the calculation of the odds ratio (OR) and its 95 % confidence interval (CI).
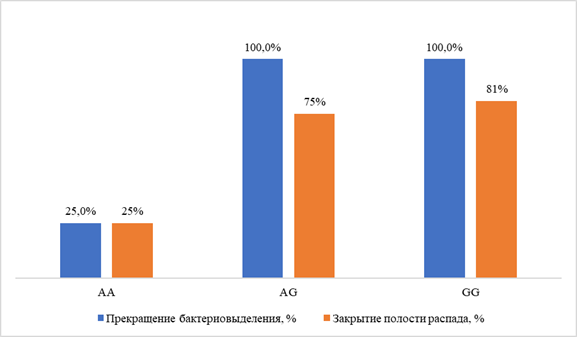
Results. Target polymorphisms were identified: SLCO1B1 (rs4149056 — in 25.8 %), ABCB1 (rs1045642 — in 75.0 %, rs2032582 — 72.2 %, rs1128503 — 77.8 %), ABCG2 (rs2231142 — in 24.3 %), CYP3A4 (rs2740574 — in 8.1 %), CYP3A5 (rs776746 — in 10.8 %). The treatment efficacy based on the criterion of cessation of bacteriosis was 89.3 % (95 % CI 72.0–97.1 %); the incidence of adverse events was 70 % (95 % CI 54.5–82.0 %), with neurotoxic reactions prevailing (in 11 of 40 patients, 27.5 %). The AA genotypes of the CYP3A5 rs776746 gene and the AA genotypes of the ABCG2 rs2231142 gene were associated with a minimum frequency of cessation of bacterial shedding: respectively, in 33 % and 0% of individuals with each variant, compared to 100% in the rest, p < 0.01; OR 0.021 (95 % CI 0.001–0.77) and 0.083 (95% CI 0.01–0.98). The risk of neurotoxic reactions was higher in the presence of the "wild" variant (genotype GG) of the ABCB1 rs2032582 gene (55.6 % vs. 16.0 % in patients with allelic polymorphisms, p = 0.034; OR 6.3; 95 % CI 1.2–33.3); gastrointestinal reactions — in the presence of the TT genotype of the ABCB1 rs1128503 gene (50.0 % vs. 10.0 %, p = 0.045; OR=9.0; 95 % CI 1.22–66.2 %).
Conclusion. Polymorphisms of CYP3A5 (rs776746, AA genotype) and ABCG2 (rs2231142, AA genotype) genes were revealed, associated with unfavorable results of treatment of patients with MDR-TB. Genetic predictors of neurotoxic and gastrointestinal reactions during treatment of patients with MDR of the pathogen were determined.
PERSONALIZED THERAPY
What is already known on this topic?
Rheumatoid arthritis (RA) is a chronic autoimmune disease, and methotrexate (MTX) is the cornerstone drug for its treatment.
The efficacy of MTX is limited: approximately 30% of patients do not respond due to insufficient effectiveness or side effects.
A major challenge is the lack of reliable tools to predict the response to MTX prior to initiating therapy.
Research is ongoing to identify genetic markers (single nucleotide polymorphisms, SNPs) associated with MTX's mechanism of action.
What does this study add?
A novel pharmacogenetic model for predicting non-response to methotrexate therapy was developed and validated.
The model incorporates a combination of five gene polymorphisms: ATIC rs2372536, MTHFR rs1801133, ADA rs244076, MTHFR rs1801131, and SLC19A1 rs1051266.
The study demonstrates that accounting for the biochemical pathways of MTX metabolism (folate and adenosine) is critical for creating an effective model, unlike a purely mathematical approach.
The model shows high sensitivity (80.2%) and stability (robust in 10/10 cross-validation).
For the model's practical application, a domestically produced test system was developed, along with software utilizing "if-then" decision rules.
How might this affect clinical practice in the foreseeable future?
The implementation of this model could enable a shift towards a personalized approach in RA treatment.
Using a genetic test, physicians could assess a patient's likelihood of responding to methotrexate before starting therapy.
This would allow for:
Promptly prescribing alternative therapies (e.g., biological agents, JAK inhibitors) for patients with a high predicted risk of non-response, avoiding a trial of ineffective MTX.
Improving overall treatment efficacy and the speed of achieving remission.
Reducing the risk of unnecessary side effects in patients unlikely to benefit from MTX.
Background. Approximately 30 % of rheumatoid arthritis (RA) patients exhibit inadequate response to methotrexate (MTX), with associated adverse effects limiting treatment efficacy, necessitating tools for predicting therapeutic outcomes [1]. The absence of robust pharmacogenetic models hinders personalized RA management.
Objective. This study aimed to develop a pharmacogenetic model to predict the risk of non-response to MTX in RA patients based on polymorphisms in genes encoding key proteins involved in MTX metabolism.
Methods. A prospective cohort study enrolled 281 RA patients meeting the European Alliance of Associations for Rheumatology criteria, receiving MTX as the initial disease-modifying antirheumatic drug. After 6 months, therapeutic response was assessed using the Disease Activity Score-28 (DAS28), identifying 170 responders and 111 non-responders. Genotyping was performed for polymorphisms in SLC19A1 (rs1051266), ABCB1 (rs1128503, rs2032582), GGH (rs3758149), FPGS (rs4451422, rs1544105), MTHFR (rs1801131, rs1801133), ATIC (rs2372536), ADA (rs244076), AMPD1 (rs17602729), ITPA (rs1127354).
Predictive models were developed using multifactor dimensionality reduction (MDR) and information analysis (Shannon entropy).
Results. The final model, incorporating five single nucleotide polymorphisms “ATIC rs2372536 + MTHFR rs1801133 + ADA rs244076 + MTHFR rs1801131 + SLC19A1 rs1051266”, achieved a sensitivity of 80.2 %, specificity of 69.4 % (OR 9.18 [95 % CI 5.19; 16.22]), and high cross-validation consistency (10/10).
Conclusion. This five-gene model demonstrates robust diagnostic performance for predicting MTX non-response in RA, with practical implementation via an “if-then” decision rule.
CASE STUDY
What is already known on this topic?
Standard Approach: Historically, cystic fibrosis (CF) treatment has been symptomatic (supportive care, enzymes, infection control).
Treatment Revolution: The advent of targeted CFTR modulators (e.g., ivacaftor, lumacaftor, tezacaftor, elexacaftor) revolutionized therapy by addressing the underlying cause of the disease.
Interaction Problem: These drugs are prone to complex drug-drug interactions via the cytochrome P450 system (CYP3A) and transporter proteins (OATP1B1, P-gp), requiring careful management.
Genetic Background: Polymorphisms in genes encoding these enzymes and transporters (e.g., CYP3A5, SLCO1B1) are known to affect the metabolism of many drugs.
What does the article add?
Proof-of-Concept Case: The article presents a specific case of a CF patient who developed hepatotoxicity and bilirubin fluctuations while on CFTR modulator therapy.
Identification of Genetic Cause: Pharmacogenetic testing identified a "problematic" genotype combination—CYP3A5 *3/*3 and SLCO1B1 *1/*5—in the patient, explaining the impaired drug metabolism and increased risk of toxicity.
Personalized Management: Based on the genetic results, a personalized management plan was developed, including avoiding specific drugs and foods and the need for dose adjustments.
How might this affect clinical practice in the foreseeable future?
Implementation of Pharmacogenetic Testing: The article justifies the feasibility of routine pharmacogenetic testing (for CYP3A5, SLCO1B1 genes) in CF patients before initiating or during issues with targeted therapy.
Improved Treatment Safety: This approach will allow for the prediction and prevention of adverse drug reactions (e.g., hepatotoxicity) before they occur, making CFTR modulator therapy safer.
Development of Personalized Regimens: Based on a patient's genetic profile, clinicians will be able to provide specific recommendations on avoiding interacting drugs and, if necessary, prescribe personalized doses from the outset, improving treatment tolerance and adherence.
Relevance. The study of pharmacokinetic and pharmacodynamic drug interactions involving components of targeted medications demonstrates the lack of available data and the significant need for research aimed at describing the likelihood, extent, and clinical impact of proposed drug interactions for individual patients and for the population of patients with cystic fibrosis.
Objective. To describe a clinical case of a patient with cystic fibrosis F508del in the CFTR gene in combination with PIDS (Common Variable Immune Deficiency) a carrier of the potentially "problematic" CYP3A5 *3/*3 and SLCO1B1 *1/*5 genotypes for liver metabolism, with an assessment of the safety of the targeted therapy for cystic fibrosis.
Materials and methods. For genetic analysis, the isolated DNA was examined using the iPLEX Pro PGx panel (Agena Bioscience) in the "VeriDose® Core Panel" modification, the patient revealed: P-glycoprotein (P-gp) gene ABCB1 (rs1045642) G/G, APOE E2/E3, CYP1A2*1A/*1F, CYP2B6*1/*1, CYP2C19*1/*1 , CYP3A4*1/*1 , CYP3A5*3/*3 , PNPLA5 (RS5764010) C/C and SLCO1B1*1/*5 .
Results. The patient's biochemical abnormalities were clarified during the selection of a drug for targeted therapy of cystic fibrosis, as well as during the use and switching of targets. Clinically insignificant abnormalities in biochemical liver function parameters were not accompanied by clinical symptoms.
Conclusion. Modern pharmacogenetic testing capabilities have made it possible to identify a potentially "problematic" combination of CYP3A *3/*3 and SLCO1B1 *1/*5 genotypes in a patient, which is associated with changes in drug metabolism in the liver. Therefore, the use of pharmacogenetic testing in patients with genetic diseases opens up opportunities for personalization and improvement of pharmacotherapy safety, allowing for the prevention or delay of organ dysfunction to enhance.
What is already known on this topic?
Definition and Causes: Vertebral artery dissection (VAD) is a tear in the artery wall with an intramural hematoma. It can be spontaneous or traumatic.
Stroke Risk: VAD is a significant cause of ischemic stroke, particularly in young adults. The stroke can occur immediately or in a delayed fashion.
Stroke Mechanisms: The primary mechanisms are arterio-arterial embolism and the propagation of thrombosis within the damaged artery.
Standard Treatment: The cornerstone of conservative therapy and secondary stroke prevention is dual antiplatelet therapy (acetylsalicylic acid + clopidogrel).
Problem of Resistance: Genetic resistance to clopidogrel, associated with polymorphisms of the CYP2C19 gene, is a known factor that reduces its efficacy.
What does the article add?
- Unique Clinical Case: It provides the first detailed description of a link between confirmed genetic resistance to clopidogrel and the development of delayed ischemic stroke specifically in a patient with VAD, despite standard therapy.
Proof of Effective Therapy Adjustment: Using a practical example, it demonstrates that replacing clopidogrel with prasugrel (which is less susceptible to genetic resistance) in such a patient is a safe and effective strategy, leading to stabilization and regression of symptoms.
Highlighting the Issue for VAD: The article emphasizes that the problem of genetic resistance to antiplatelets in VAD is understudied and requires more attention.
How might this affect clinical practice in the foreseeable future?
- Changing Approach to "Breakthrough" Strokes: The occurrence of a stroke in a VAD patient while on standard antiplatelet therapy may become an indication for testing for genetic resistance to clopidogrel.
Therapy Personalization: If resistance is identified, practice may shift towards early replacement of clopidogrel with alternative drugs (prasugrel, ticagrelor) to prevent recurrent vascular events.
Stimulating Research: The article points to the need for finding simple and accessible laboratory markers (e.g., within thromboelastometry) to help rapidly identify VAD patients who may not respond to clopidogrel, without the need for complex genetic testing.
A clinical case of a 35-year-old man with dissection of the left vertebral artery is presented, in connection with which dual antiplatelet therapy in the form of aspirin and clopidogrel, as well as anticoagulant therapy with enoxaparin, was prescribed to prevent the development of thromboembolic complications.
On day 5, the patient developed numbness in the right extremities, dysphagia and dysarthria, increased ataxia, left-sided ptosis, right-sided hemiparesis and hemihypesthesia. A control MRI scan of the brain revealed a focus of ischemia in the medulla oblongata. Pharmacogenetic testing was performed with the study of genetic resistance to antiplatelet drugs with the determination of polymorphic variants rs4244285*2, rs4986893*3, rs12248560*17 of the CYP2C19 gene. It was revealed that the patient was a carrier of the CT genotype according to the rs12248560 polymorphic variant, the GA genotype according to the rs4244285 polymorphic variant, and the GG genotype according to the rs4986893 polymorphic variant of the CYP2C19 gene. This corresponds to a variant of an intermediate metabolizer with an indistinctly defined effect of clopidogrel. The above clinical observation with the development of delayed ischemic stroke after spinal artery dissection (DPA) draws attention to the problem of genetic resistance to antiplatelet agents in this patient population. The development of delayed ischemic stroke in DPA is the basis for determining genetic resistance to antiplatelet agents and the subsequent possible change in treatment tactics.
ISSN 2686-8849 (Online)